10+ Calculate_Qc_Metrics
Datasc_example_counts datasc_example_cell_info pd. After I use function adata scread_visium count_fileV1_Human_Lymph_Node_filtered_feature_bc_matrixh5.
Python Kallisto Bustools
FastQC Is a very commonly used NGS QC.

. Convert annotation from GenomicRanges to Simple Annotation. Stack Exchange Network Stack Exchange network consists of 181 QA communities. On the starting nucleic acids After Library preparation Post-Sequencing Ultimately the best QC of your NGS experiments is.
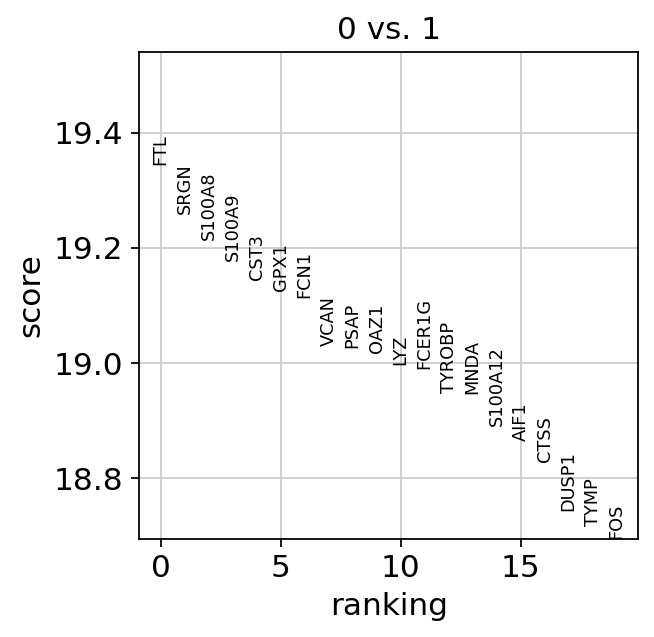
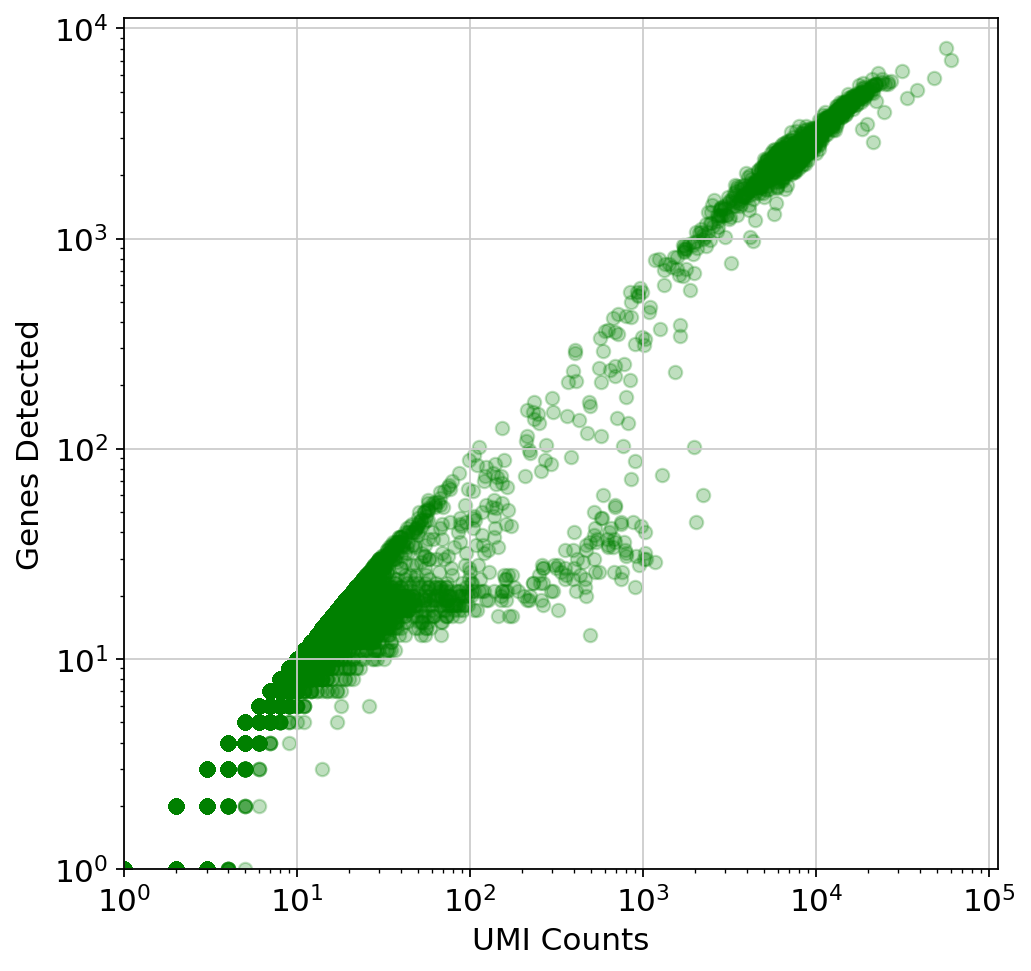
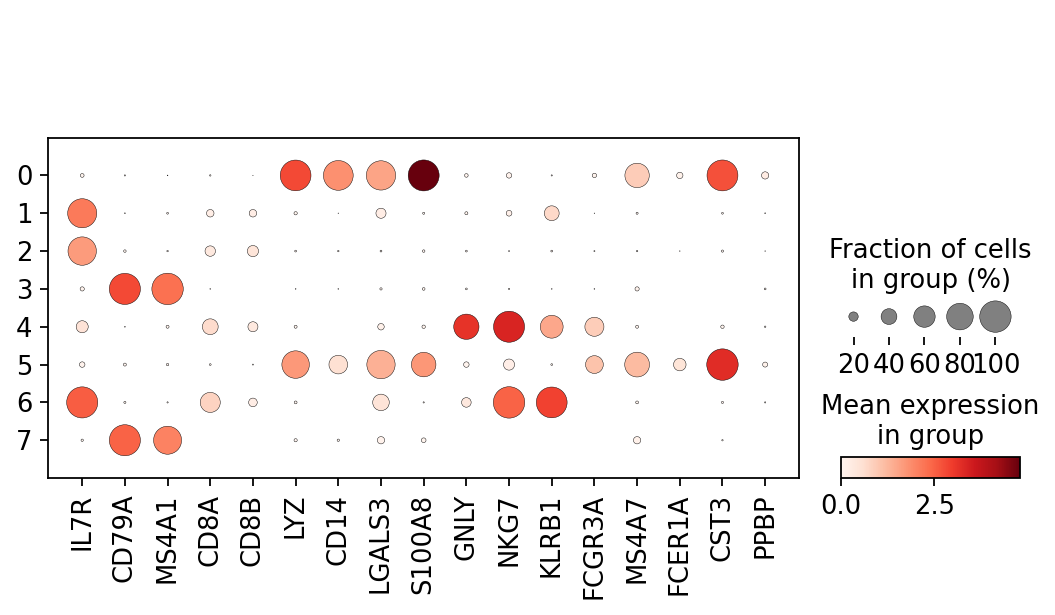
We can for example calculate the percentage of mitocondrial and ribosomal. Here is what the multiqc report should look like we can view. Number_of_genes number of genes detected.
One containing quality control metrics about cells and one containing metrics about genes. Quality Control QC metrics can be viewed in QC reports and in final reports. They use calculateQCMetric function to calculate the quality metrics but I am gettin.
31 rows Quality Control QC metrics. Cd workspacealign mkdir post_align_qc cd post_align_qc multiqc workspacealign tree. There are 3 main areas where QC can be applied to NGS.
Analysis of NGS quality scores A quality score is an estimate of the probability of that base being called wrongly q -10 x log 10 p. The QC statistics added are. I was also surprised since this should be the first few functions people run.
This function is housed in the. The calculate_qc_metrics function returns two dataframes. With this workflow well-known single-sample QC metrics and additional metrics specific for tumor-normal pairs can be calculated.
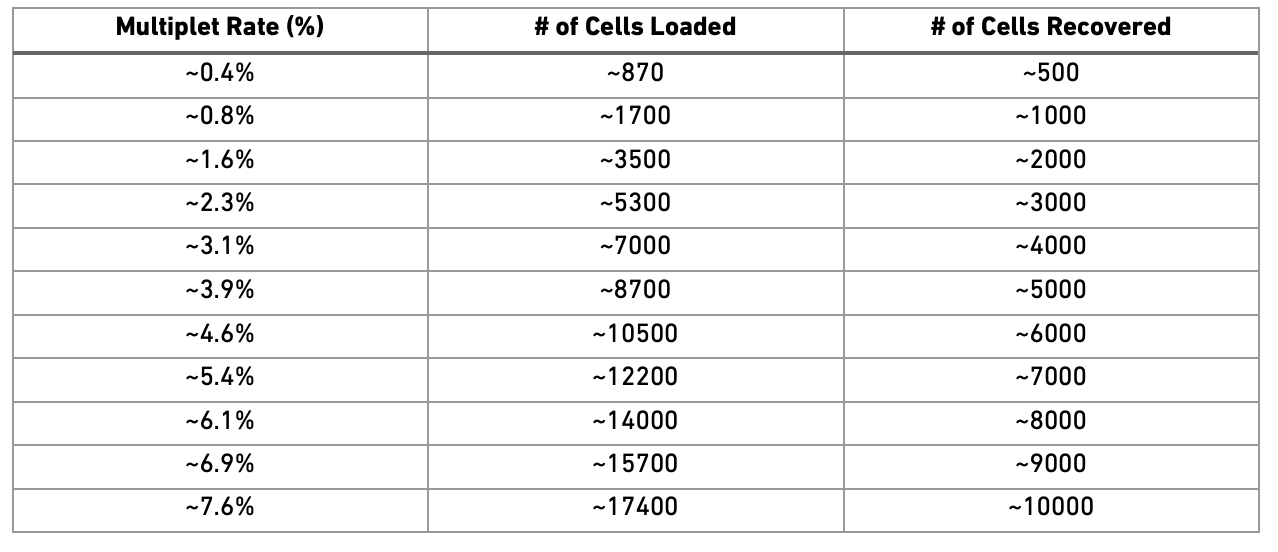
For planning purposes the Patient Risk Sigmas used for the calculation of run sizes are 528. The segmentation into different tools offers a. Get QC metrics using gene count matrix.
Calculate QC metrics from gene count matrix. Run MultiQC to produce a final report. Import gene annotation anno_to_saf.
This function calculate basic QC metrics with scanpyppcalculate_qc_metrics. Having the data in a suitable format we can start calculating some quality metrics. The metrics are based on data that are included in an analysis.
Total_count_per_cell sum of read number after UMI deduplication. The calculator results are shown below. Additionally total counts and the ratio of unspliced and spliced matrices as well as the cell cycle scores.
The calculated sigmas are 528 909 and 939 resp. Calculate_qc_metrics was there for a long time.

沈德咏 Keithlevina
Python Kallisto Bustools
Python Kallisto Bustools

Scanpy 01 Qc

沈德咏 Keithlevina

Scanpy 01 Qc

The Benchmarking Dataset Open Problems In Single Cell Analysis
Sc Pp Calculate Qc Metrics Runtime Error Issue 1147 Scverse Scanpy Github
Scanpy 01 Qc
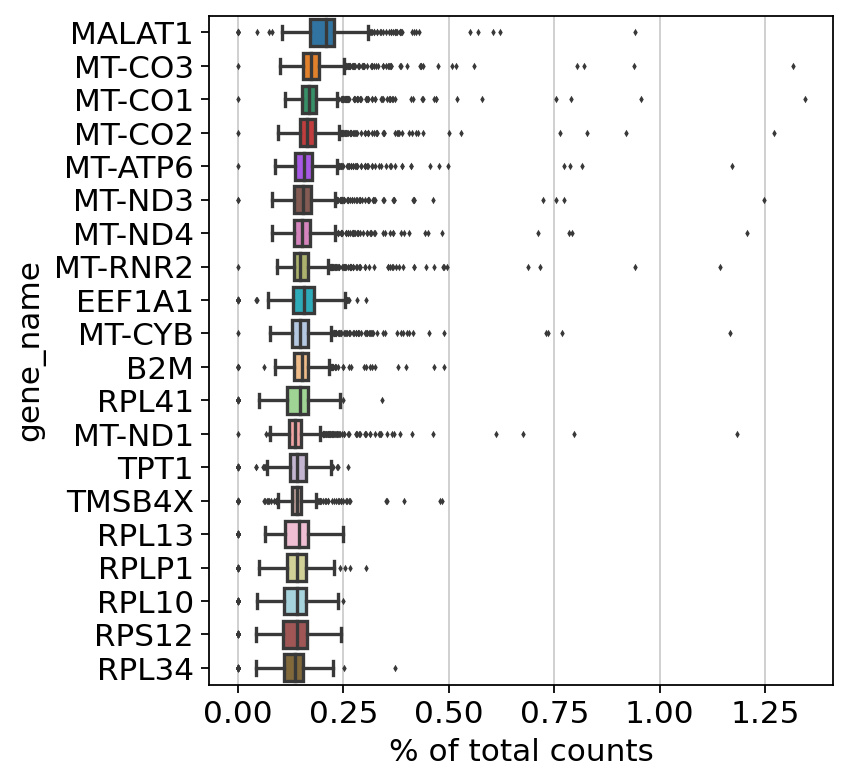
Python Kallisto Bustools
Scanpy Sc Pp Calculate Qc Metrics Adata Qc Vars Mt Inplace True Issue 1341 Scverse Scanpy Github

Scanpy 01 Qc

沈德咏 Keithlevina
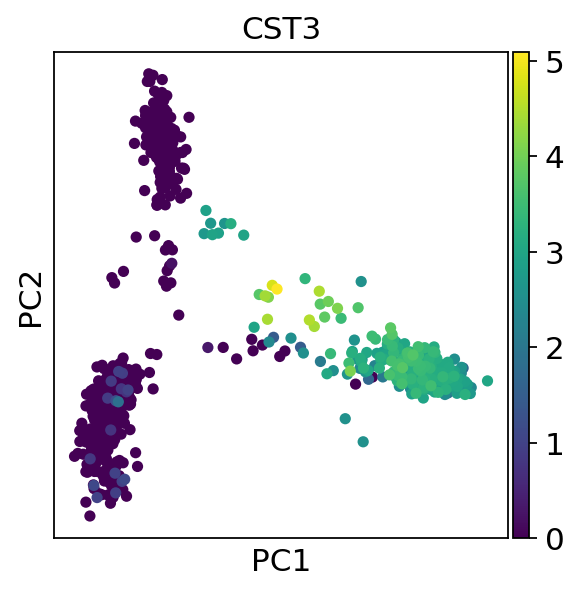
Python Kallisto Bustools
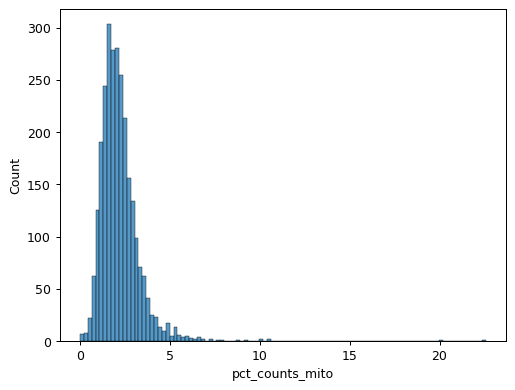
Scanpy Pp Calculate Qc Metrics Scanpy 1 9 1 Documentation
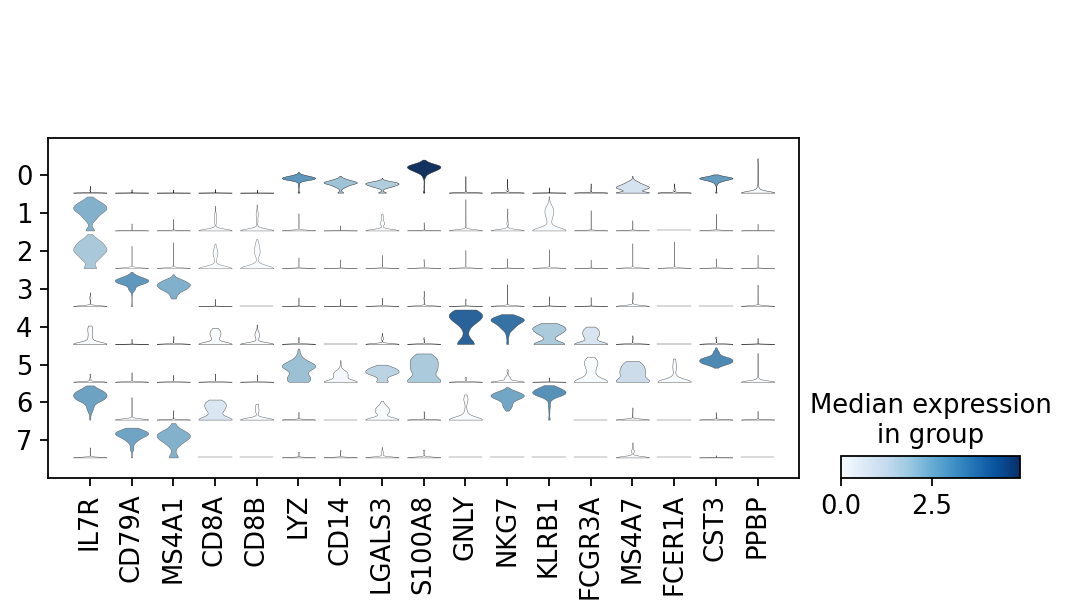
Python Kallisto Bustools

Scanpy 01 Qc
Komentar
Posting Komentar